- Home
- low coverage
- Efficient phasing and imputation of low-coverage sequencing data using large reference panels
Efficient phasing and imputation of low-coverage sequencing data using large reference panels
5 (668) · $ 23.99 · In stock

Ultra Low-Coverage Whole-Genome Sequencing as an Alternative to Genotyping Arrays in Genome-Wide Association Studies. - Abstract - Europe PMC
Frontiers Comparison of Genotype Imputation for SNP Array and Low-Coverage Whole-Genome Sequencing Data
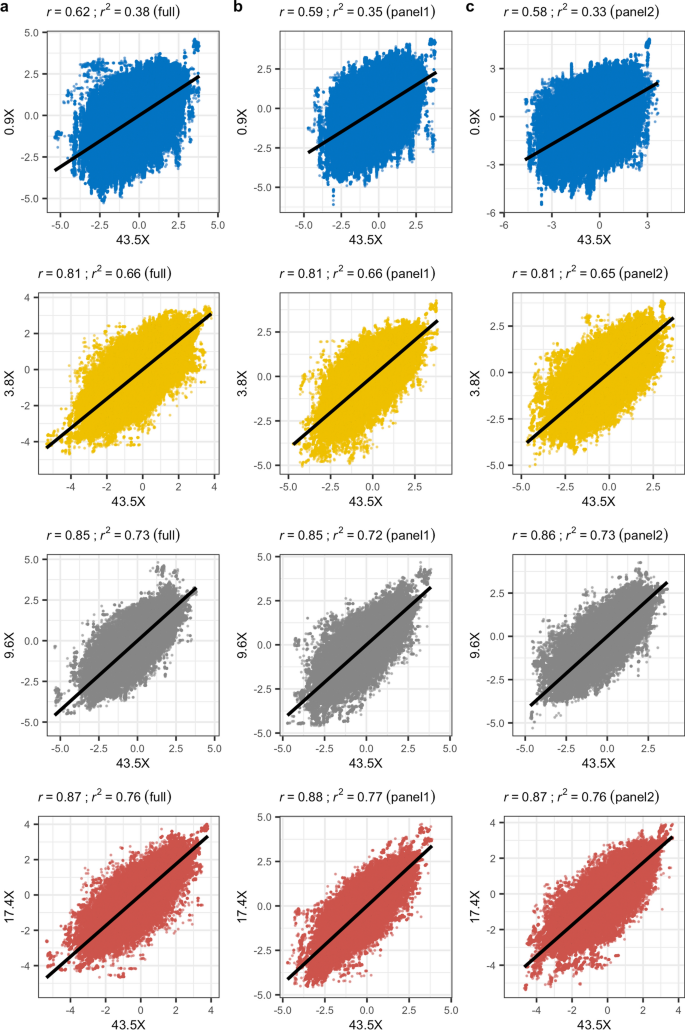
A cautionary tale of low-pass sequencing and imputation with respect to haplotype accuracy, Genetics Selection Evolution

A population-specific reference panel for improved genotype imputation in African Americans

Efficient phasing and imputation of low-coverage sequencing data using large reference panels
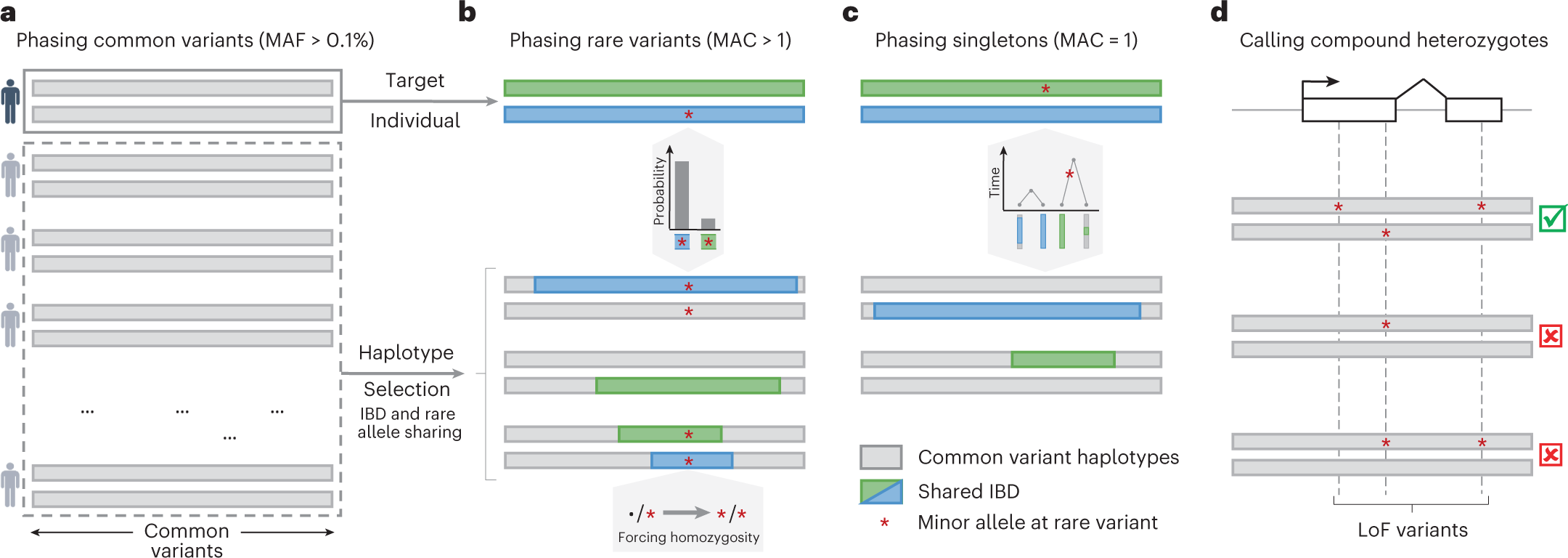
Accurate rare variant phasing of whole-genome and whole-exome sequencing data in the UK Biobank

PDF) Efficient phasing and imputation of low-coverage sequencing data using large reference panels

SEAD: an augmented reference panel with 22,134 haplotypes boosts the rare variants imputation and GWAS analysis in Asian population

Performance and accuracy evaluation of reference panels for genotype imputation in sub-Saharan African populations - ScienceDirect

PDF] GeneImp: Fast Imputation to Large Reference Panels Using Genotype Likelihoods from Ultralow Coverage Sequencing
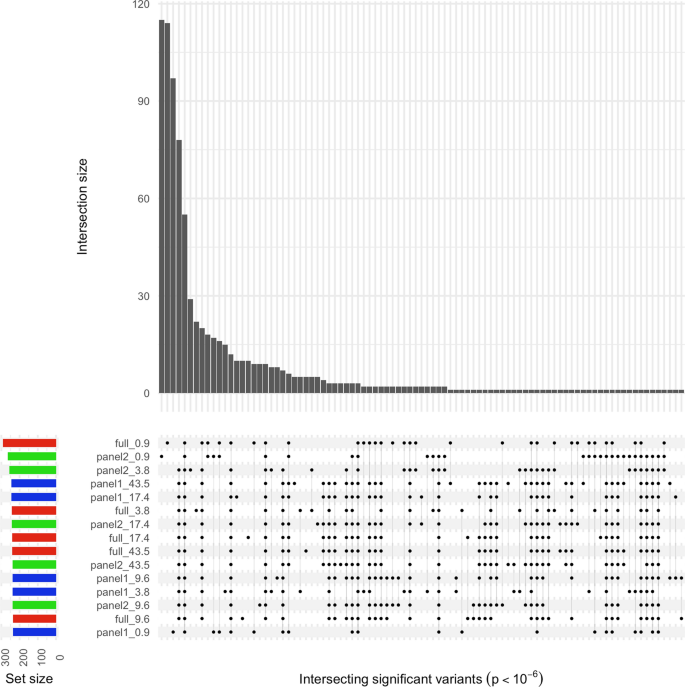
A cautionary tale of low-pass sequencing and imputation with respect to haplotype accuracy, Genetics Selection Evolution

PDF] GeneImp: Fast Imputation to Large Reference Panels Using Genotype Likelihoods from Ultralow Coverage Sequencing